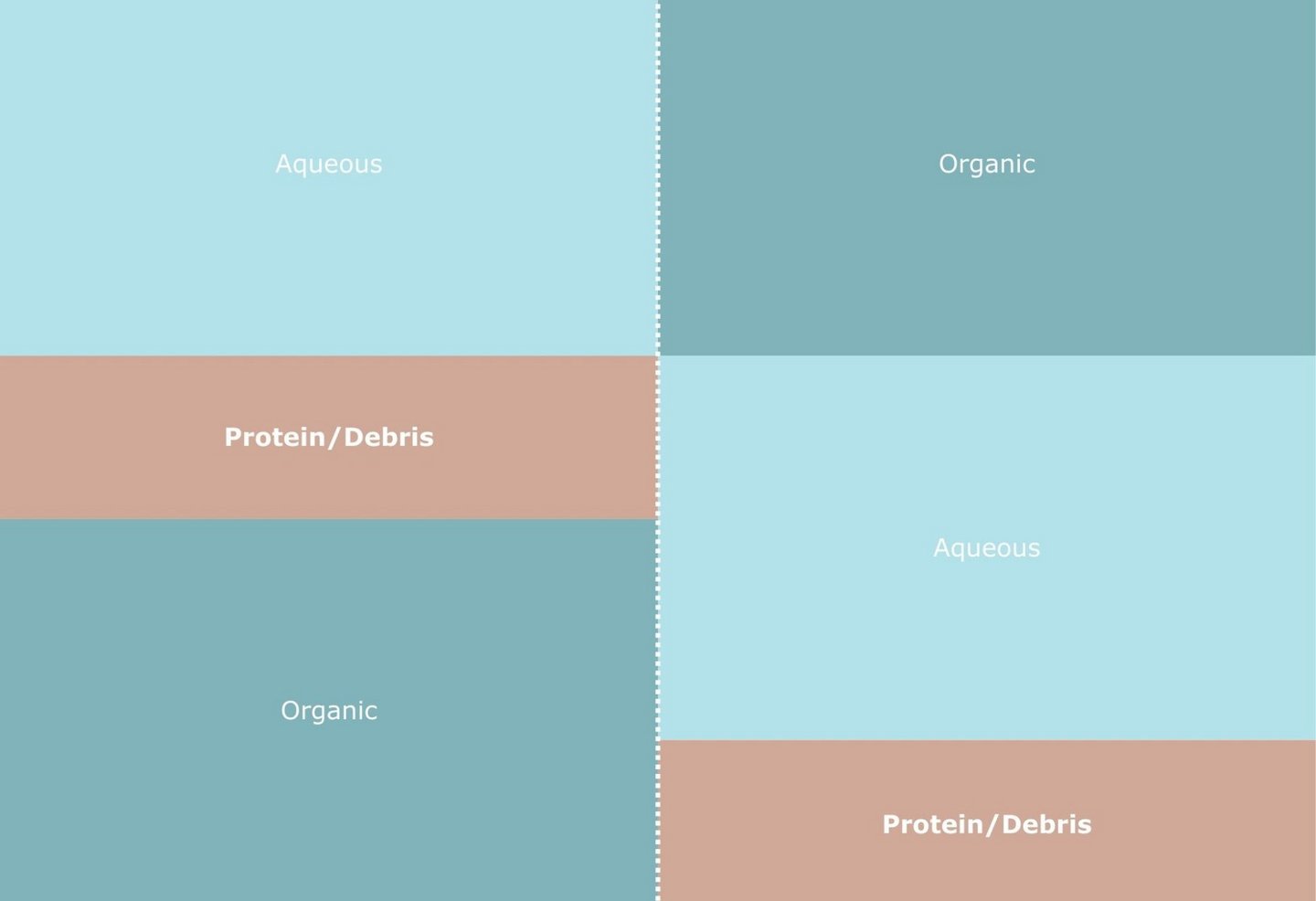
As we balance speed and depth, the definition of "depth" continues to expand. While the single-sample workflow integrates three layers (Metabolome, Lipidome, Proteome), new research pushes the boundaries even further.
The analytical landscape is diversifying. We now have MAE-MTBE for rapid, green lipid extraction (Bellinghieri, Campaniello) and Cold MTBE-SP3 for deep, comprehensive multi-omics (Gegner).
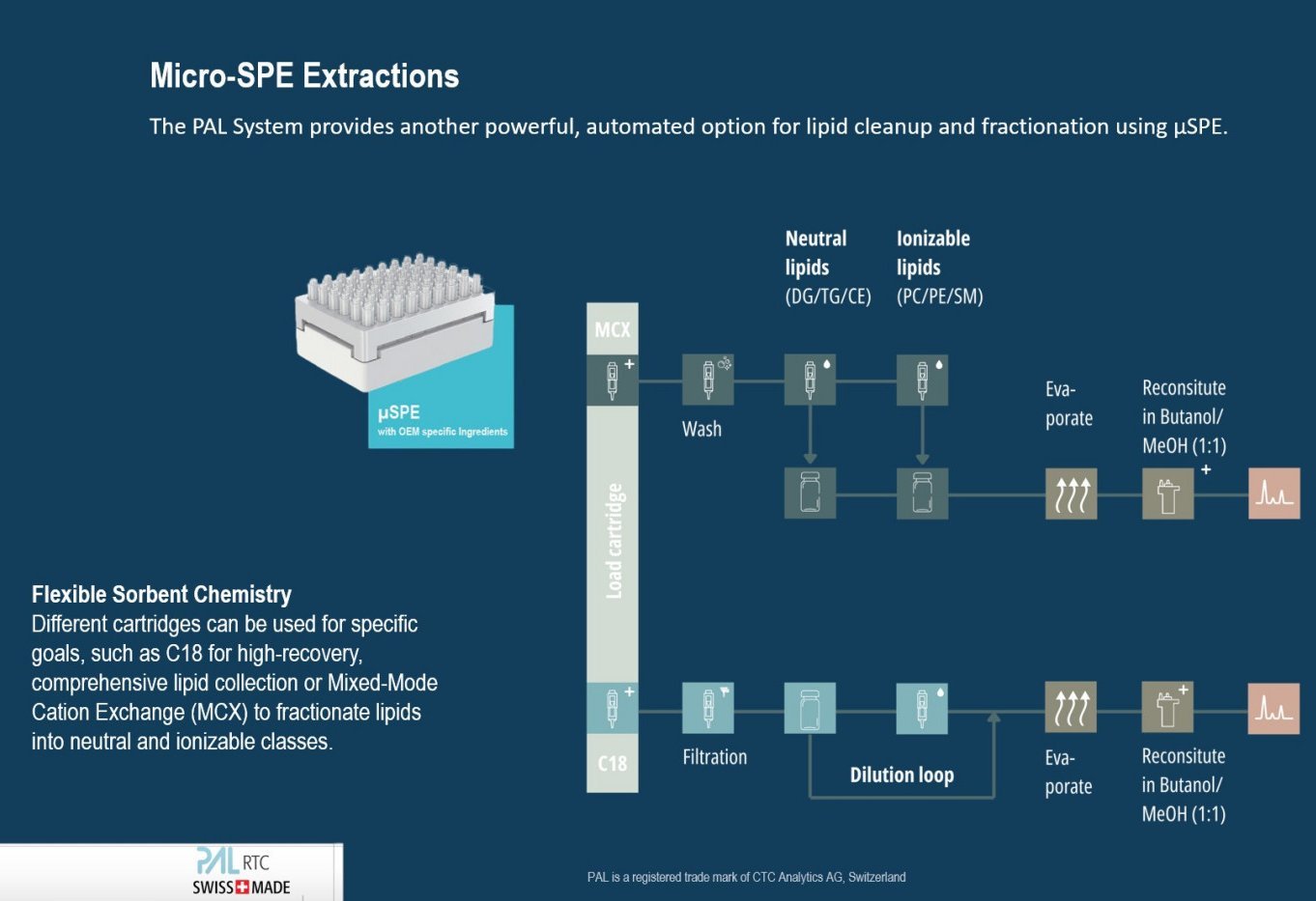
The common denominator is the need for precision. Whether you are injecting a microwave-extracted lipid sample or fractionating a complex multi-omics extract via Micro-SPE, the reproducibility of the result depends on the reproducibility of the handling. By automating these distinct workflows on a PAL System, laboratories can flexibly switch between "Green Speed" and "Deep Biology" without compromising on data integrity. Even smaller automation processes, such as adding internal standards or dilutions/calibration curves, will improve the results noticeably.
-
Bellinghieri, C., Giacoppo, G., Schincaglia, A., Ferrara, D., Purcaro, G., Cavazzini, A., Pasti, L., Stevanin, C., Chenet, T., Marchetti, N., Maietti, A., Franchina, F.A., & Beccaria, M. (2026). "Applicability of MTBE-based lipid extraction methods assisted by microwave in food analysis. Statistical comparison and greenness evaluation with Soxhlet and Matyash." Talanta, 299, 129124.
-
Campaniello, M., Nardelli, V., Zianni, R., Tomaiuolo, M., Miedico, O., Iammarino, M., & Mentana, A. (2024). "Microwave-Assisted Extraction/UHPLC-Q-Orbitrap-MS-Based Lipidomic Workflow for Comprehensive Study of Lipids in Soft Cheese." Foods, 13(7), 1033. doi.org/10.3390/foods13071033
-
Cavalluzzi, M.M., Lamonaca, A., Rotondo, N.P., Miniero, D.V., Muraglia, M., Gabriele, P., Corbo, F., De Palma, A., Budriesi, R., De Angelis, E., Monaci, L., & Lentini, G. (2022). "Microwave-Assisted Extraction of Bioactive Compounds from Lentil Wastes: Antioxidant Activity Evaluation and Metabolomic Characterization." Molecules, 27(21), 7471. doi.org/10.3390/molecules27217471
-
Gegner, H.M., Naake, T., Aljakouch, K., Dugourd, A., Kliewer, G., Müller, T., Schilling, D., Schneider, M.A., Rohrbach, N.K., Grünewald, T.G.P., Hell, R., Rodriguez, J.S., Huber, W., Poschet, G., & Krijgsveld, J. (2024). "A single-sample workflow for joint metabolomic and proteomic analysis of clinical specimens." Molecular Systems Biology, 20, 248–278.
-
Hu, Q., He, Y., Wang, F., Wu, J., Ci, Z., Chen, L., Xu, R., Yang, M., Lin, J., Han, L., & Zhang, D. (2021). "Microwave technology: a novel approach to the transformation of natural metabolites." Chinese Medicine, 16, 87. doi.org/10.1186/s13020-021-00500-8
-
Marques, M.S., Sussulini, A., Ferreira, P.C., & Pereira, T.F.D. (2022). "The Importance of Sample Preparation for Omics Analysis: Which Extraction Method is the Most Suited for my Biological Question?" Brazilian Journal of Analytical Chemistry, 10(40), 99-111.
-
Li, W., Sun, J., Sun, R., Wei, Y., Zheng, J., Zhu, Y., & Guo, T. (2025). "Integral-Omics: Serial Extraction and Profiling of Metabolome, Lipidome, Genome, Transcriptome, Whole Proteome and Phosphoproteome Using Biopsy Tissue." Analytical Chemistry, 97(2), 1190-1198.