For many years, the process from collecting a biological sample, whether it is blood, plasma, or tissue, to getting meaningful data has involved several detailed steps. Scientists are very familiar with established sample preparation techniques like liquid-liquid extraction (LLE), where analytes are partitioned between two immiscible liquids [2, 3], and solid phase extraction (SPE), where analytes are isolated based on their affinity to a solid sorbent material [2, 3]. While these methods are standard in the lab, they can be quite demanding in terms of time, resources, the amount of (often organic) solvents used, and even the quantity of sample needed [1, 4, 5, 6].
While modern mass spectrometers offer incredible sensitivity, the field of metabolomics, in particular, operates on the principle "garbage in, garbage out." Without effective sample preparation to remove interfering components (like salts and proteins) and to enrich target analytes, even the most advanced analytical instrument cannot deliver reliable or meaningful biological insights. This need has driven the development of greener and more solvent-efficient techniques, such as Micro-SPE (Micro Solid Phase Extraction), which aim to reduce waste and sample size while still offering effective clean-up. However, when scientists are working with very small or precious samples, or trying to detect substances present in tiny amounts, the challenges remain, and the quest for more efficient, reliable, and affordable approaches, particularly those that can be miniaturized and automated, is a constant in the scientific community [3].
The Leiden team's innovative solution focuses on a technique called electro-extraction (EE). Unlike LLE or SPE, which often rely on passive diffusion or chemical affinity, EE actively uses an electric field to gently guide charged target molecules out of a complex sample and into a clean solution, ready for analysis [2]. This active transport can significantly increase extraction speed and efficiency [2]. This method is known for its ability to concentrate analytes effectively while using minimal amounts of solvents and sample, addressing some of the key limitations of more traditional approaches [1, 2, 7, 8]. EE is particularly promising for handling small sample volumes and for high-throughput analysis, making it an attractive option for modern metabolomics [3, 9].
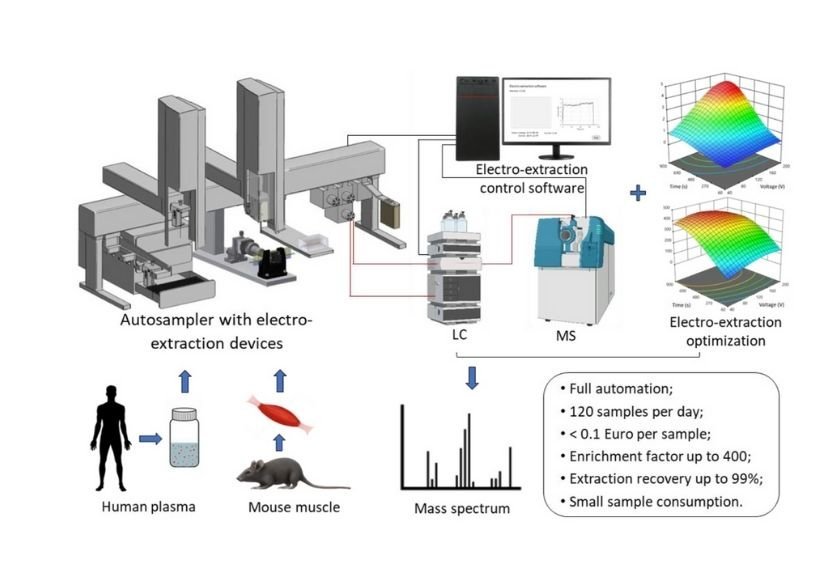
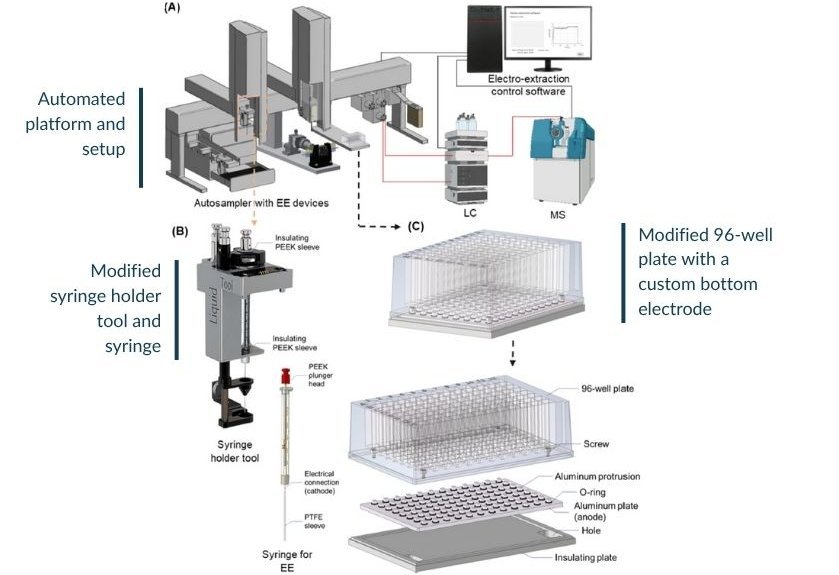
What makes this new work by Dr. He and colleagues particularly exciting is the level of automation achieved. They have combined a PAL System (Dualhead) with custom-designed 96-well plates featuring a built-in electrode, all connected to an LC/MS setup (Liquid Chromatography/Mass Spectrometry) for detailed analysis [1]. This is not just about speeding up one step; it is about automating the entire workflow from sample introduction to data acquisition.
The system's performance, as detailed in the paper and its supplementary information, is impressive. The researchers optimized various parameters, such as voltages and extraction times, using a Design of Experiment (DoE) approach to get the best possible results for a range of acylcarnitines. The supplementary data in He's publication (Table S2, S3, S4 and Figure S4 from [1]) illustrate this careful optimization process, showing how different conditions affect the enrichment of these molecules.
The outcomes of this work are compelling: the system can achieve an enrichment of target molecules by up to 400 times, with nearly all of the desired analytes (up to 99% recovery) being captured [1]. This means scientists can detect even very low levels of analytes. Furthermore, the automated setup can process between 96 and 120 samples in a single day, at a remarkably low cost of less than 0.1 Euro per sample [1]. This high throughput and cost-effectiveness are intriguing for larger studies or routine labs.
The true value of a new scientific tool is demonstrated when it is applied to real biological questions. Dr. He's automated EE platform has been rigorously tested with human plasma samples, showing excellent sensitivity and reproducibility (as detailed in Table 1 of his publication [1] and Figure S5 in the supplemental material, which shows enrichment factors in plasma).
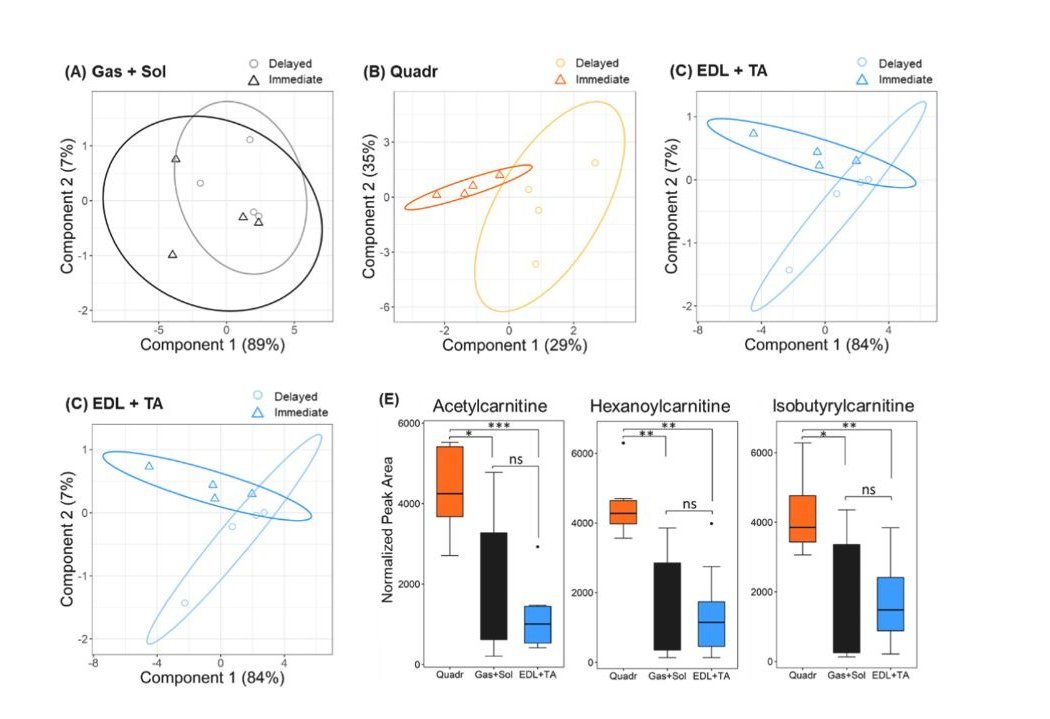
Beyond plasma, the system was also used to study mouse muscle tissues, providing insights relevant to sarcopenia, an age-related condition characterized by loss of muscle mass and function [1, 11, 12]. The research explored how quickly muscle samples need to be processed after collection to ensure the stability of acylcarnitines. The findings, supported by data in Figure 5 of the main paper [1] and Table S8 of its supplement, indicated that a 15-minute delay at room temperature before freezing the tissue did not significantly alter the acylcarnitine levels in various muscle types. This has practical implications for researchers in designing their sample collection protocols.
The study also shed light on how acylcarnitine levels differ across various muscle types in a mouse model of accelerated aging (Ercc1Δ/− mice), which typically yield very small tissue samples [1]. For example, the quadriceps muscle generally showed higher acylcarnitine levels compared to other muscle groups like the gastrocnemius and soleus combined [1, Figure 5E, Figure S7 from the supplement of [1]]. These observations contribute to a better understanding of muscle metabolism in the context of aging and disease.
This leap in automated sample preparation is more than a technical feat, it is a powerful enabler for a wide spectrum of scientific inquiry. By dramatically reducing the manual effort, time, and cost involved, such systems empower researchers to design and execute more ambitious studies. Simultaneously, it allows routine labs to offer more reliable and cost-effective services, making it more accessible to a broader range of scientists. This is especially vital for discovering new biomarkers for diseases, advancing clinical research, and realizing the promise of personalized medicine, all of which often rely on analyzing large numbers of samples with high precision. The ability to reliably work with small, precious samples also opens up new research avenues that might have been previously out of reach.
[1] He, Y., Miggiels, P., Harms, A., Rijksen, Y., Brandt, R. M. C., Vermeij, W. P., ... & Hankemeier, T. (2025). A fully automated, high-throughput electro-extraction and analysis workflow for acylcarnitines in human plasma and mouse muscle tissues. Analytica Chimica Acta, 1364, 344224.
[2] Oedit, A., Ramautar, R., Hankemeier, T., & Lindenburg, P. W. (2016). Electroextraction and electromembrane extraction: advances in hyphenation to analytical techniques. Electrophoresis, 37(9), 1170-1186.
[3] Miggiels, P., Wouters, B., van Westen, G. J. P., Dubbelman, A. C., & Hankemeier, T. (2019). Novel technologies for metabolomics: More for less. Trends in Analytical Chemistry, 120, 115323.
[4] Henion, J., Brewer, E., & Rule, G. (1998). Sample preparation for LC/MS/MS: analyzing biological and environmental samples. Analytical Chemistry, 70(19), 650a-656a.
[5] Xu, R. N., Fan, L., Rieser, M. J., & El-Shourbagy, T. A. (2007). Recent advances in high-throughput quantitative bioanalysis by LC–MS/MS. Journal of Pharmaceutical and Biomedical Analysis, 44(2), 342-355.
[6] Raterink, R. J., Lindenburg, P. W., Vreeken, R. J., Ramautar, R., & Hankemeier, T. (2014). Recent developments in sample-pretreatment techniques for mass spectrometry-based metabolomics. TrAC Trends in Analytical Chemistry, 61, 157-167.
[7] Song, A. Y., & Yang, J. (2019). Efficient determination of amphetamine and methylamphetamine in human urine using electro-enhanced single-drop microextraction with in-drop derivatization and gas chromatography. Analytica Chimica Acta, 1045, 162-168.
[8] Lindenburg, P. W., Ramautar, R., & Hankemeier, T. (2013). The potential of electrophoretic sample pretreatment techniques and new instrumentation for bioanalysis, with a focus on peptidomics and metabolomics. Bioanalysis, 5(22), 2785-2801.
[9] Oedit, A., Hankemeier, T., & Lindenburg, P. W. (2021). On-line coupling of two-phase microelectroextraction to capillary electrophoresis - Mass spectrometry for metabolomics analyses. Microchemical Journal, 162, 105741.
[10] Fielding, R. A., Vellas, B., Evans, W. J., Bhasin, S., Morley, J. E., Newman, A. B., ... & Zamboni, M. (2011). Sarcopenia: an undiagnosed condition in older adults. Current consensus definition: prevalence, etiology, and consequences. International working group on sarcopenia. Journal of the American Medical Directors Association, 12(4), 249-256.
[11] Santilli, V., Bernetti, A., Mangone, M., & Paoloni, M. (2014). Clinical definition of sarcopenia. Clinical Cases in Mineral and Bone Metabolism, 11(3), 177.